IDAA
Indel Detection by Amplicon Analysis
IDAA Rationale
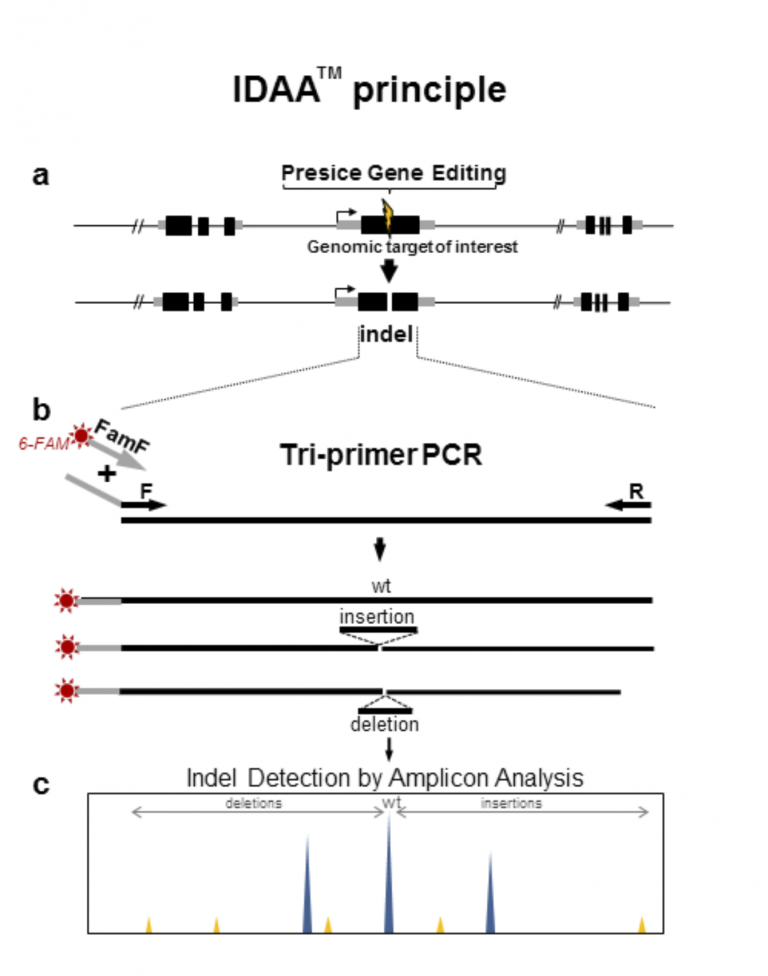
IDAATM is a superior method for identification of insertion and deletions (indels) induced by precise gene editing¹. These editing events generate double-stranded DNA breaks at a desired target site, which stimulate the cells natural DNA repair pathways to homologous recombination and non homologous end joining that lead to a variety of rearrangements at the breakpoint2. Traditional ways of indel identification include; i) enzyme mismatch cleavage assays, which provide for insufficient identification of the induced indel’s and ii) next generation DNA sequencing, which is costly, time and labor intensive, and poorly suited for high throughput screening of hundreds or thousands of clones often required to select for desirable multi-allelic editing events that often occur at low frequency.
IDAA™ is a novel strategy that combines use of a simple amplicon labelling strategy with the high throughput capability of DNA fragment analysis by automated Capillary Electrophoresis³. IDAA™ is suitable for detecting indels in both cell pools with low efficiency targeting, single sorted cells and is ideally suited for high throughput detection of indels down single base events, estimation of “cutting” efficiencies of targeting tools, and evaluation of off-target events at candidate loci¹.
[1] Yang, 2., Steentoft, C, Hauge, C., Hansen, L., Lind-Thomsen, A., Nicola, F., Vester-Christensen, M.B., Frödin, M., Clausen, H., Wandall, H.H., and Bennett, E.P. (2015) Fast and Sensitive Detection of Indels Induced by Precise Gene Targeting. Nucleic Acids Research, doi: 10.1093/nar/gkv126.
[2] Steentoft C., Bennett E.P., Schjoldager K.T., Vakhrushev S.Y., Wandall H.H., Clausen H. (2014) Precision genome editing: a small revolution for glycobiology. Glycobiology. Aug 24(8):663-80
[3] Andersen, P.S., Jespersgaard, C., Vuust, J., Christiansen, M. and Larsen, L.A. (2003) Capillary electrophoresis-based single strand DNA conformation analysis in high-throughput mutation screening. Hum. Mutat, 21, 455-65.
[4] Duda, K., Lonowski, La, Kofoed-Nielsen, M., Ibarra, A., Delay, C.M., Kang, Q., Yang, Z., Pruett-Miller, S.M., Bennett, E.P., Wandall, H.H., et al. (2014) High-efficiency genome editing via 2A-coupled co expression of fluorescent proteins and zinc finger nucleases or CRISPR/Cas9 nickase pairs. Nucleic Acids Res., 2014 Jun;42(10):84
IDAA Methodology*
IDAATM is based on a single step tri-primer PCR setup with a universal 6-FAM 5′-labelled primer, designed to be specific for an extension placed on one of the target specific primers, thus enabling one-step fluorophore labelling of amplicons derived from any given target¹ (see illustration). The tri-primer amplification assay has been standardized for general use in detecting a broad range of targets in a multitude of species. Following target amplification, the fluorophore labelled amplicons can easily be detected with great sensitivity and with accurate size evaluation using standard DNA fragment analysis by capillary electrophoresis methodology2. The whole methodology is outlined above.
*Patent application covering parts of the methodology pending
[1] Yang, Z., Steentoft, C, Hauge, C., Hansen, L., Lind-Thomsen, A., Nicola, F., Vester-Christensen, M.B., Frödin, M., Clausen, H., Wandall, H.H., and Bennett, E.P. (2015) Fast and Sensitive Detection of Indels Induced by Precise Gene Targeting. Nucleic Acids Research, doi: 10.1093/nar/gkv126.
[2] Andersen, P.S., Jespersgaard, C., Vuust, J., Christiansen, M. and Larsen, L.A. (2003) Capillary electrophoresis-based single strand DNA conformation analysis in high-throughput mutation screening. Hum. Mutat., 21, 455–65.
How to order your IDAATM kit
If you have already designed your primers you can go to our order page HERE or contact us at oligo@tagc.com.
When you place your order we will make sure that your Forward primer is 5′-modified by addition of the 5′-extension AND we will supply you with the 6-Fam modified probe (there is no need to order the 6-Fam probe separately since it is already included in the Kit). Also you will receive the entire IDAA Kit at our discounted price – you save more than 25% compared to list prices of the individual components.
If you have not yet designed your primers, we recommend that you use the free Primer3 database.
For best results with the IDAA Kit we recommend the following changes to the default settings in the Primer3 database:
- Primer Size should be: Min 21, Opt 23, Max 25
- Primer TM should be: Min 57, Opt 60, Max 63
- Product Size Ranges should be 200-500
All other settings could be default.
When you have your primer sequences ready please go to our order page HERE or contact us at oligo@tagc.com
The IDAA Kit contains:
- 1x Reverse primer, RPC purified (your design)
- 1x Forward primer, RPC purified (your design + a 5′-extension)
- 1x 6-Fam probe, HPLC purified (IDAA design)
- 1x Shipping box (users guide is supplied upon IDAA-kit order)
Price for the entire IDAA kit is only DKK 350 / USD 50 / EUR 47
The delivered amount is enough for approx. 1.000 reactions (equivalent to a synthesis scale of 40nmol)
Turnaround time is two days plus shipping.